|
Engineer novel synthetic cells Natural cells are modified for broad applications by exploiting their ability to synthesize biomolecules, respond and adapt to environments, and change their structures dynamically. But, it remains challenging to engineer synthetic cells that are robust, safe, and effective, akin to microrobots. My lab integrates material and genetic approaches to create novel synthetic cells (ranging from bacterial pathogens to human primary cells) that respond to environmental stimuli. Our work will generate novel synthetic cells that are safe and effective for environmental remediation, cancer therapy, antipathogen treatment, and microbiome applications.
Representative publications |
|
|
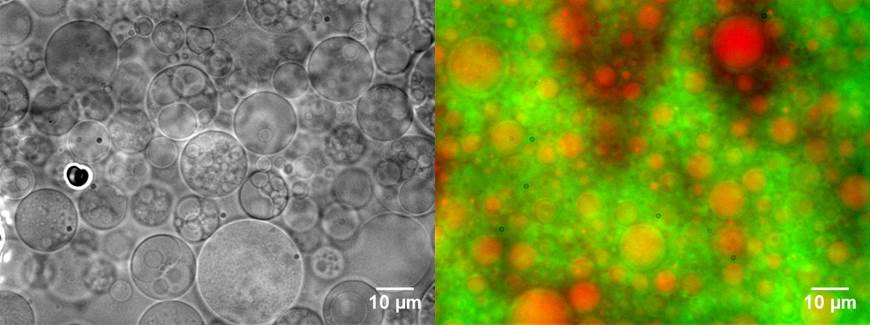
Engineer novel synthetic vesicles Natural cells secrete vesicles that are composed of phospholipids and biomolecules. Harnessing and mimicking these vesicles could create novel solutions for biomedical applications. But, creating synthetic vesicles that are multifunctional and “smart” remains challenging. My lab builds synthetic vesicles that mimic specific properties of living cells. We use synthetic biology approaches that integrate advanced cloning methods, proteomics, high-resolution imaging, and cell-free systems. The synthetic vesicles are minimal and functionalized with proteins, genes, and biomaterials. Our work will generate novel synthetic vesicles for disease diagnostic and treatment.
Representative publications |
|
|
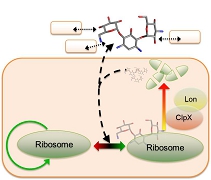
Understand and control the dynamics of synthetic biological networks Biological networks consist of biomolecules linked by various feedback loops. These feedback loops operate under noisy cellular environments and can cause emergent behavior of cells. Understanding and controlling the flow of information through complex biological networks are crucial for the engineering of synthetic cells and vesicles. My lab uses mathematical modeling and quantitative measurements to reveal emergent dynamics of synthetic biological networks. We are among the first to discover the holistic interactions between genetic and non-genetic factors, including molecular crowding, antagonistic signaling pathways, and host-circuit interactions. Our work will create quantitative frameworks to predict and control the functions of synthetic biological networks.
Representative publications |
|
|
Develop high-throughput synthetic biology platforms Much of life science revolves around understanding and exploiting the function of proteins. Yet, only a small subset of proteins is routinely studied in basic research or used in applications. My lab integrates cell-free protein synthesis, molecular tools, microfluidics, and computational algorithms to accelerate the study of proteins. Our work will enable the high-throughput study of “difficult” and understudied proteins.
Representative publications |
|
Our toolbox
| Real-time microscopy 
|
High-throughput assay 
|
Proteomics 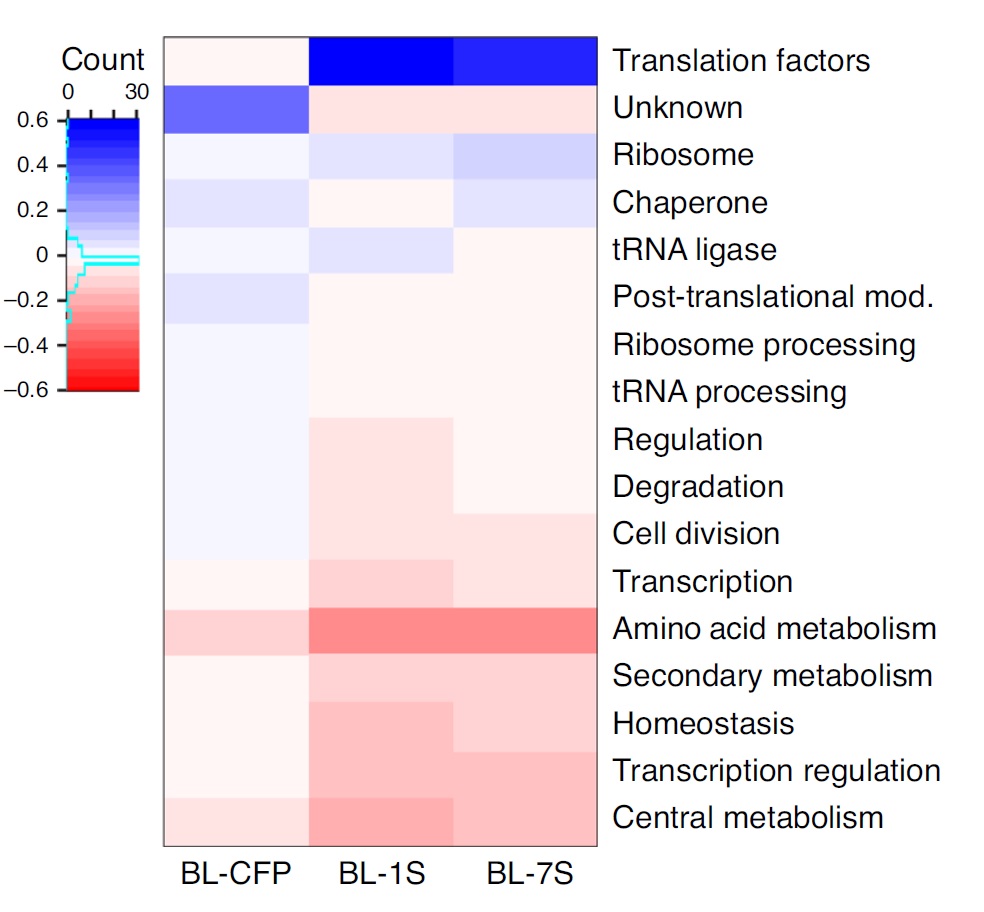
|
High-throughput cloning and CRISPR-Cas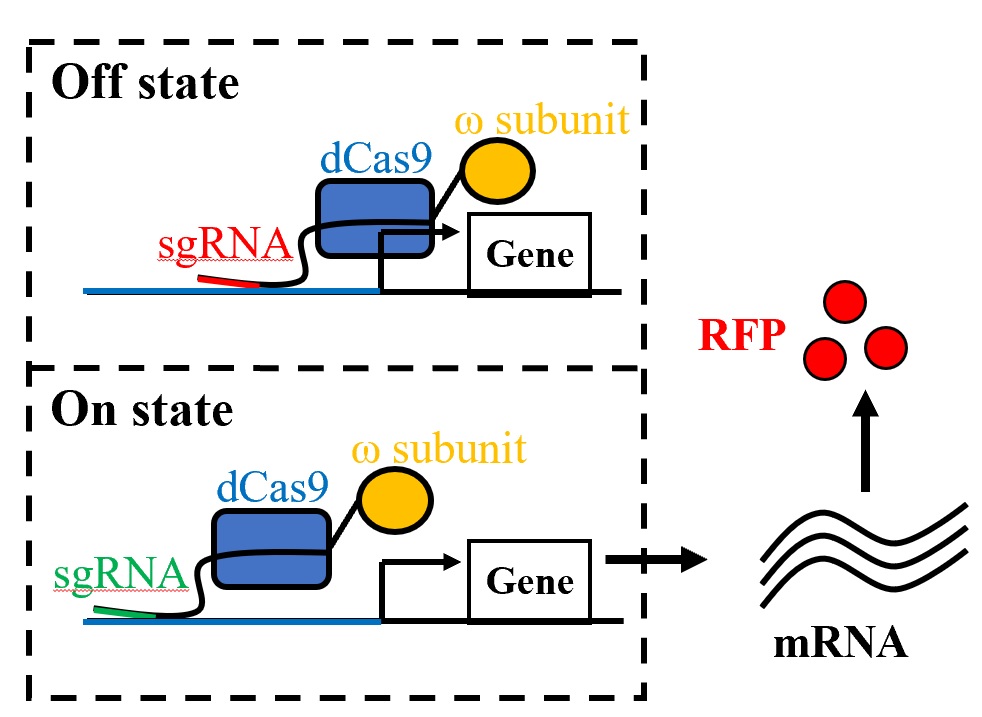 |
Artificial cells & cell-free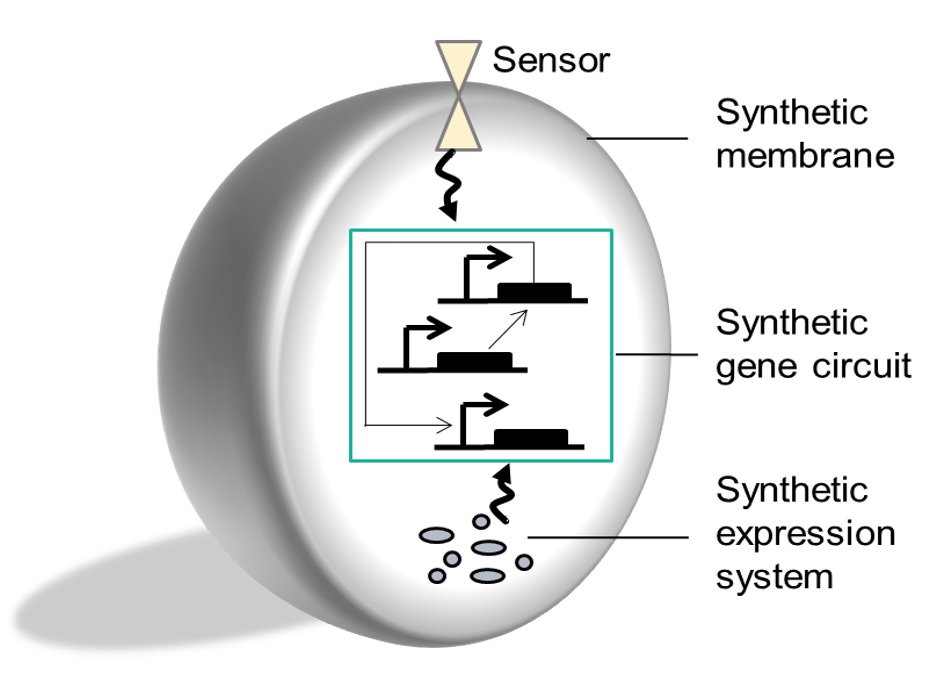 |
Bacteria and Mammalian Cells  |
Robotics |
Matlab, C++, Python |
High-performance computing; Machine learning |

